Projects
WEPP: Wastewater-Based Epidemiology using Phylogenetic Placements
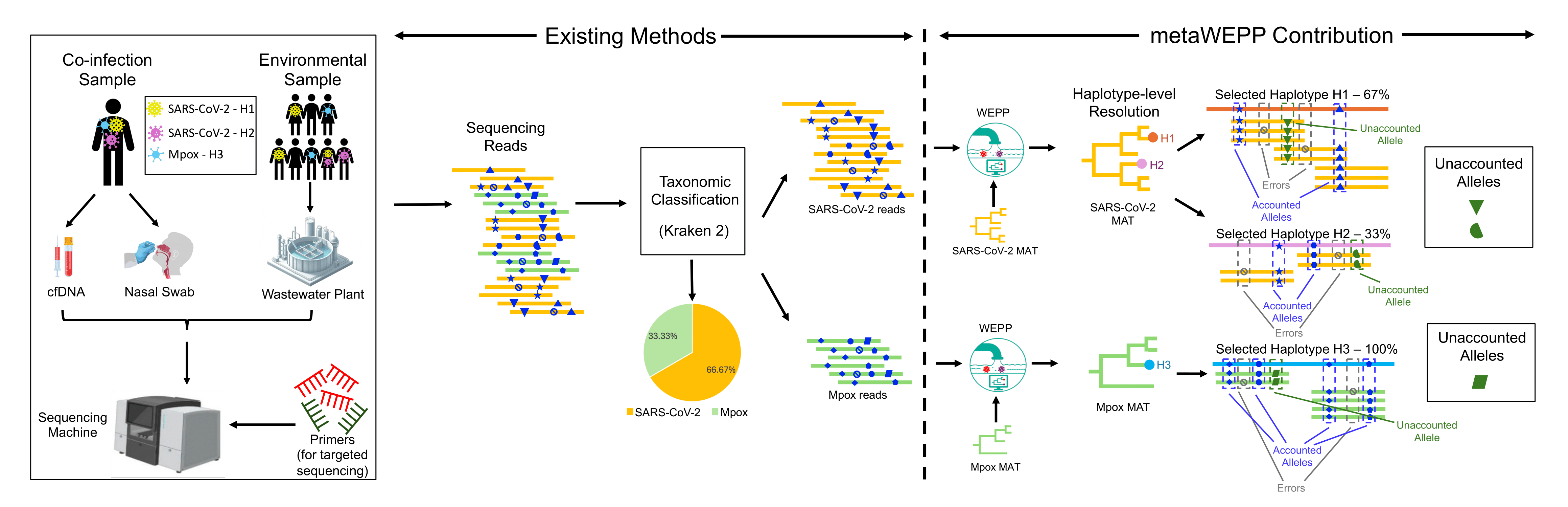
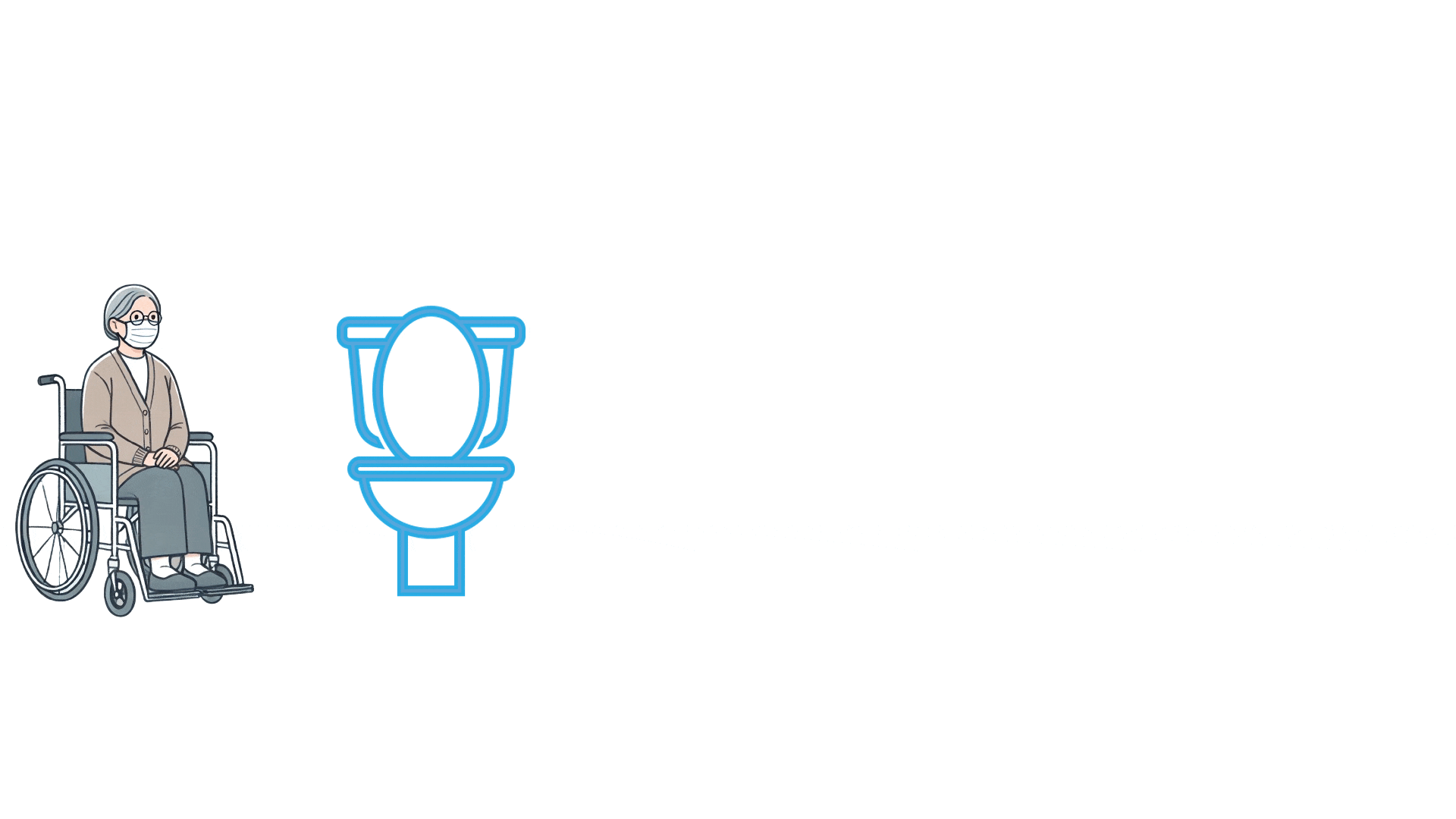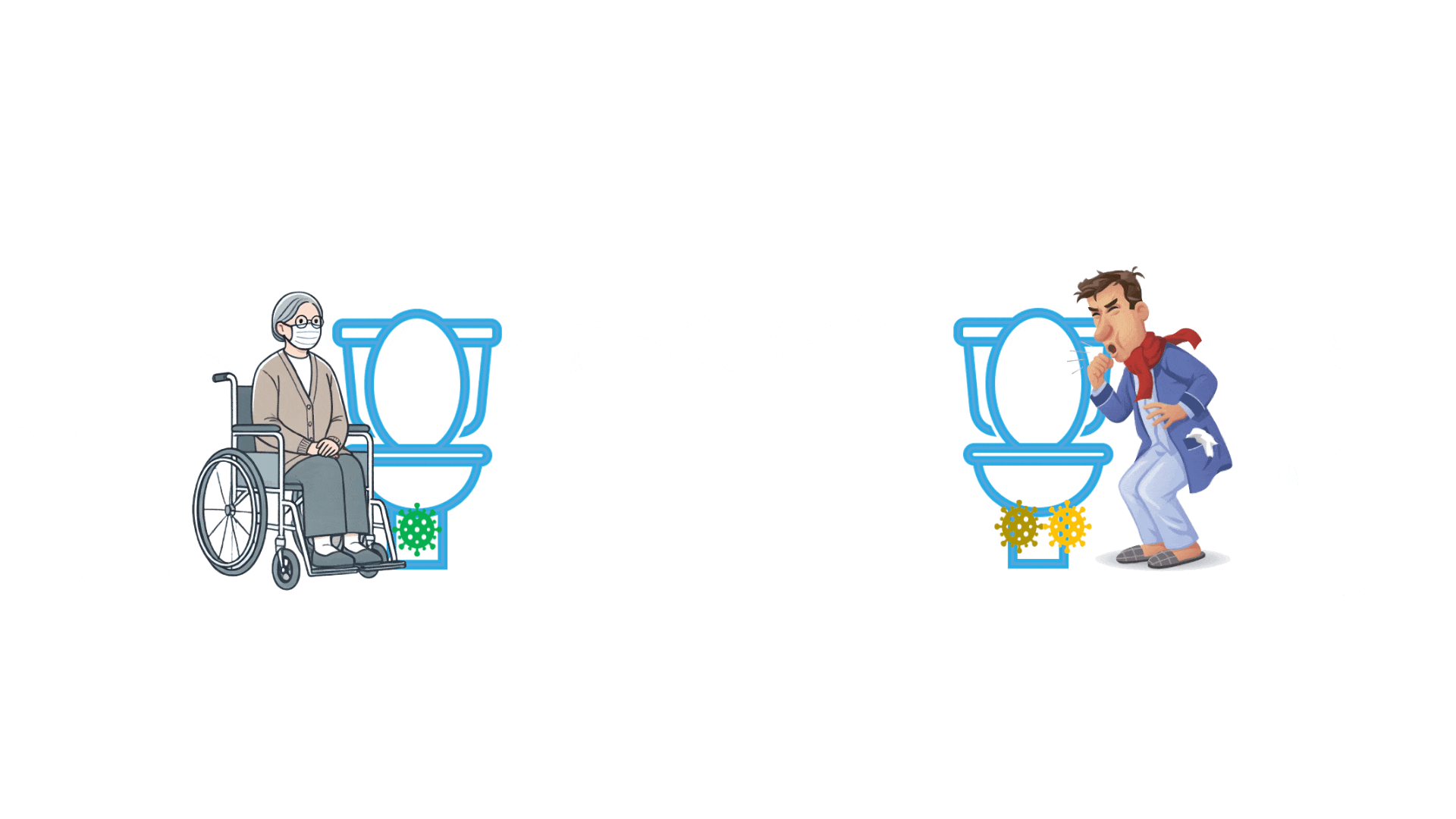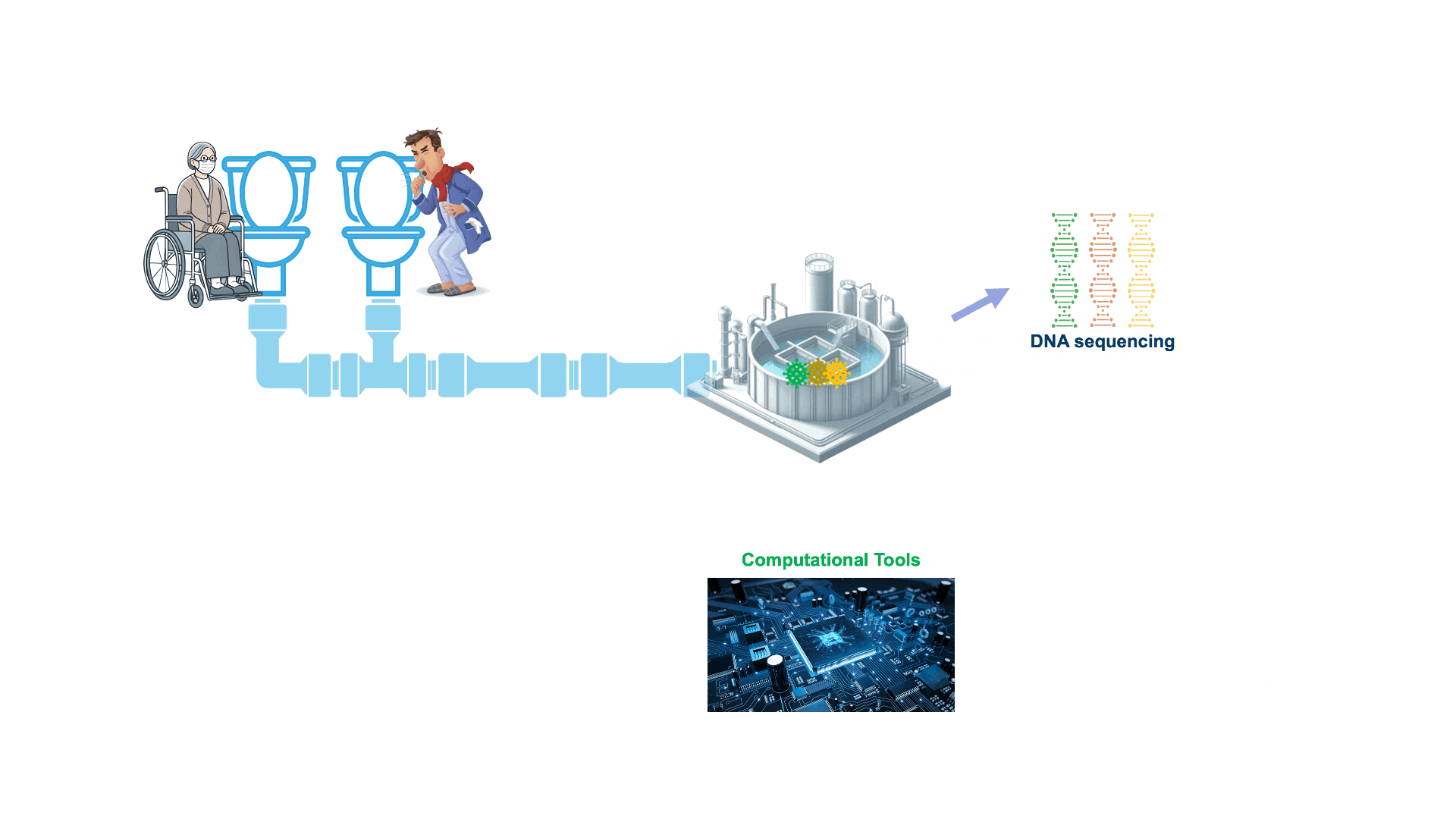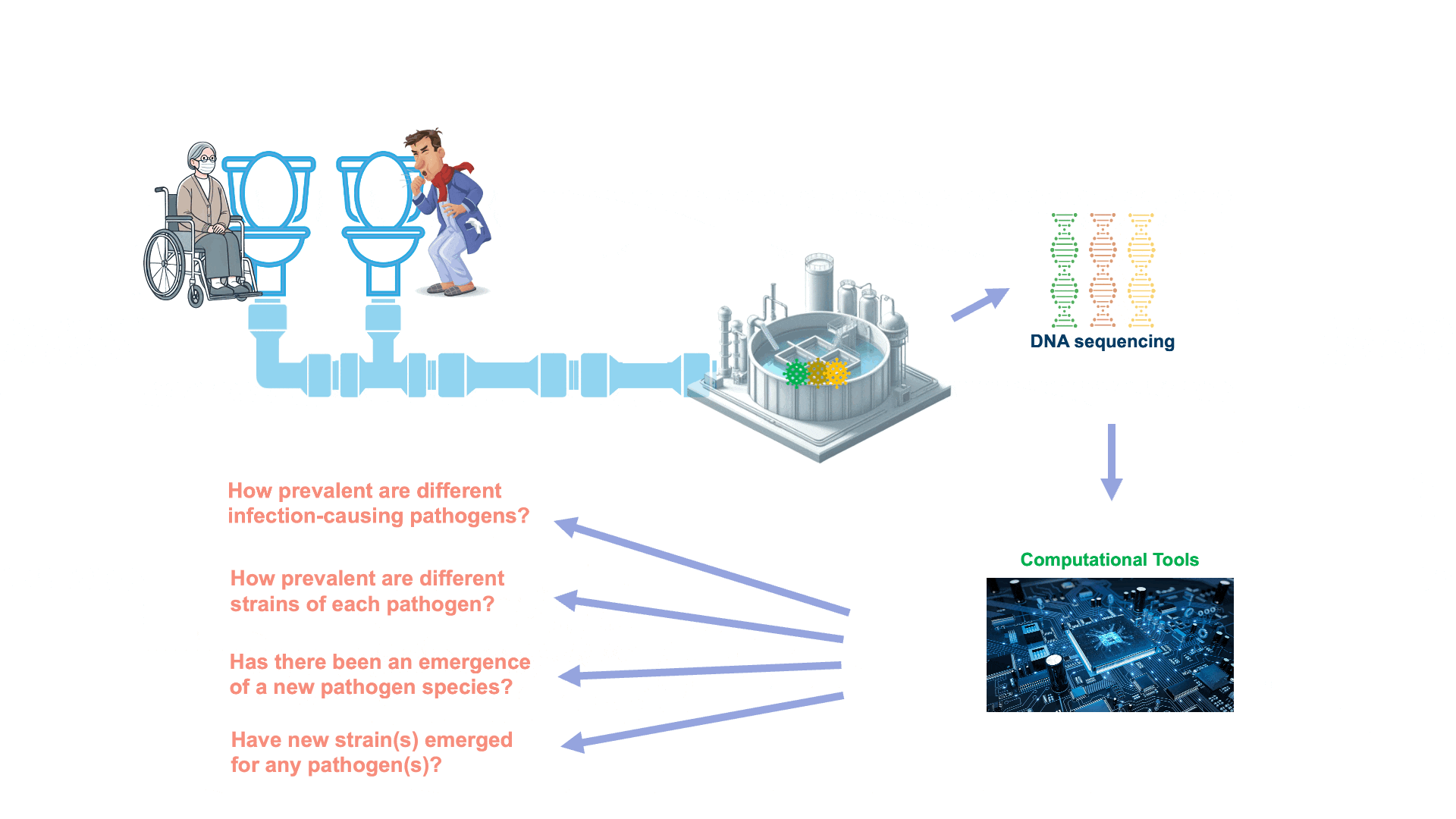
Wastewater contains a treasure trove of public health information, including genetic traces of viruses and bacteria circulating in a community. However, this data is highly complex — more like a puzzle with millions of mixed-up pieces. WEPP is a powerful tool designed to solve this puzzle far more effectively than existing methods.
WEPP analyzes small fragments of genetic material (sequencing reads) from wastewater and accurately places them onto a pathogen’s “family tree,” known as a phylogenetic tree. By doing so, WEPP identifies the specific nodes in the tree, corresponding to unique genome sequences, from which these fragments most likely originated. This higher resolution allows public health officials to not only determine which pathogen variants are present in a community, but also to identify emerging strains before they receive an official designation. Such early detection is critical for monitoring outbreaks and enabling timely, effective public health responses.
metaWEPP: Improving resolution of metagenomic analysis using WEPP
While WEPP is designed to detect specific pathogens in wastewater, metaWEPP extends this approach to complex environmental samples—such as soil, water, air, and bodily fluids like cell-free DNA—that contain genetic material from many different organisms. Rather than a single puzzle, these samples resemble thousands of puzzles mixed together.
metaWEPP efficiently untangles this complexity by first identifying which organism each genetic fragment belongs to, and then applying WEPP to identify the specific genome variants for each organism. This enables a detailed view of the microbial composition of a sample, supporting applications ranging from clinical diagnostics to environmental and ecological monitoring.